Reuters donosi, że While Russia has confirmed the world's fourth largest tally of coronavirus cases, it has a relatively low death toll from the associated disease, COVID-19[...] But data released by the Rosstat State Statistics Service on Sept. 4 show there were 57,800 excess deaths between May and July, the peak of the outbreak. The figure was calculated by comparing fatalities over those three months in 2020 with the average number of May-July deaths between 2015 and 2019. The excess total is more than three times greater than the official May-July COVID-19 death toll of 15,955. Co mnie zmotywowało do poszukania danych źródłowych.
Okazało się że dość prosto jest je pobrać ze strony Urzędu Statystycznego Federacji Rosyjskiej (https://eng.gks.ru/ a konkretnie https://showdata.gks.ru/finder/) Ponieważ okazało się to aż tak proste, to pobrałem nie tylko zgony ale także urodzenia. W okresie 2015--2020 (miesięcznie).
Dane są w formacie XSLX. Kolumny to miesiące wiersze poszczególne regiony i ogółem Federacja. Eksportuję do CSV używając LibreOffice i wybieram wiersz z danymi dla całej federacji. W rezultacie mam trzy wiersze: rok-miesiąc, urodzenia i zgony. Obracam moim super skryptem do transpozycji plików CSV:
month;urodzenia;zgony 2015-01-01;149269.99;174722.99 2015-02-01;144591.99;156737 2015-03-01;160974.99;175809.99 ...
Trzy wykresy liniowe przedstawiające dynamikę zgonów, urodzin i przyrostu naturalnego (czyli różnicy między urodzinami a zgonami)
library("dplyr")
library("ggplot2")
library("scales")
spanV <- 0.5
## przyrost naturalny
d <- read.csv("UZRT.csv", sep = ';', header=T, na.string="NA");
d$diff <- d$urodzenia - d$zgony
pz <- ggplot(d, aes(x= as.Date(month), y=zgony)) +
geom_line(color="steelblue", size=.6, alpha=.5) +
geom_point(color="steelblue", size=.8) +
## Trend
geom_smooth(method="loess", se=F, span=spanV, color="red3") +
## Skala na osi X-ów
scale_x_date( labels = date_format("%y/%m"), breaks = "4 months") +
xlab(label="") +
ylab(label="deaths") +
geom_vline(xintercept = as.Date("2015-07-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2016-07-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2017-07-01"), alpha=.25, size=1)+
geom_vline(xintercept = as.Date("2018-07-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2019-07-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2020-07-01"), alpha=.25, size=1) +
labs(caption="https://showdata.gks.ru/finder/") +
ggtitle("Russian Federation: Deaths (ths)",
subtitle= sprintf("red line: loess with span=%.2f ; gray vertical: july", spanV))
pu <- ggplot(d, aes(x= as.Date(month), y=urodzenia)) +
geom_line(color="steelblue", size=.6, alpha=.5) +
geom_point(color="steelblue", size=.8) +
geom_smooth(method="loess", se=F, span=spanV, color="red3") +
scale_x_date( labels = date_format("%y/%m"), breaks = "4 months") +
xlab(label="") +
ylab(label="births") +
labs(caption="https://showdata.gks.ru/finder/") +
geom_vline(xintercept = as.Date("2015-08-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2016-08-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2017-08-01"), alpha=.25, size=1)+
geom_vline(xintercept = as.Date("2018-08-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2019-08-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2020-08-01"), alpha=.25, size=1) +
ggtitle("Russian Federation: Births (ths)",
subtitle= sprintf("red line: loess with span=%.2f ; gray vertical: august", spanV))
pp <- ggplot(d, aes(x= as.Date(month), y=diff)) +
geom_line(color="steelblue", size=.6, alpha=.5) +
geom_point(color="steelblue", size=.8) +
geom_smooth(method="loess", se=F, span=spanV, color="red3") +
scale_x_date( labels = date_format("%y/%m"), breaks = "4 months") +
geom_vline(xintercept = as.Date("2015-05-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2016-05-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2017-05-01"), alpha=.25, size=1)+
geom_vline(xintercept = as.Date("2018-05-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2019-05-01"), alpha=.25, size=1) +
geom_vline(xintercept = as.Date("2020-05-01"), alpha=.25, size=1) +
xlab(label="") +
ylab(label="balance") +
labs(caption="https://showdata.gks.ru/finder/") +
ggtitle("Russian Federation: natural balance (ths)",
subtitle= sprintf("red line: loess with span=%.2f ; gray vertical: may", spanV))
Wykres liniowy dla miesięcy jednoimiennych (MJ):
library(ggplot2)
library(dplyr)
library(tidyr)
library(scales)
options(dplyr.print_max = 1e9)
size0 <- 1.2
size1 <- 0.6
size2 <- 0.6
tt <- read.csv("UZRT.csv", sep = ';', header=T, na.string="NA");
tt$rok <- substr(tt$month, 1, 4)
tt$yyyymmdd <- as.Date(sprintf ("%s-%s", substr(tt$month, 6, 7),
substr(tt$month, 9, 10) ), format="%m-%d")
## Obliczenie przyrostu naturalnego
tt$diff <- tt$urodzenia - tt$zgony
## Obliczenie
tt.yy2020 <- tt %>% filter(rok==2020) %>% as.data.frame
tt.yy2019 <- tt %>% filter(rok==2019) %>% as.data.frame
tt.yy2018 <- tt %>% filter(rok==2018) %>% as.data.frame
tt.yy2017 <- tt %>% filter(rok==2017) %>% as.data.frame
tt.yy2016 <- tt %>% filter(rok==2016) %>% as.data.frame
tt.yy2015 <- tt %>% filter(rok==2015) %>% as.data.frame
pf <- ggplot() +
ggtitle("Russian Federation: births 2015--2020") +
geom_line( data = tt.yy2020, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2020'), alpha=.5, size=size0) +
geom_point( data = tt.yy2020, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2020'), alpha=.5, size=size0) +
geom_line( data = tt.yy2019, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2019'), alpha=.5, size=size1) +
geom_point( data = tt.yy2019, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2019'), alpha=.5, size=size0) +
geom_line( data = tt.yy2018, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2018'), alpha=.5, size=size1) +
geom_point( data = tt.yy2018, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2018'), alpha=.5, size=size1) +
geom_line( data = tt.yy2017, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2017'), alpha=.5, size=size1) +
geom_point( data = tt.yy2017, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2017'), alpha=.5, size=size1) +
###
geom_line( data = tt.yy2016, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2016'), alpha=.5, size=size1) +
geom_point( data = tt.yy2016, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2016'), alpha=.5, size=size1) +
geom_line( data = tt.yy2015, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2015'), alpha=.5, size=size2) +
geom_point( data = tt.yy2015, mapping = aes(x=yyyymmdd, y = urodzenia, colour = '2015'), alpha=.5, size=size1) +
scale_x_date( labels = date_format("%y/%m"), breaks = "2 months") +
labs(caption="https://showdata.gks.ru/finder/") +
ylab(label="ths") +
xlab(label="") +
labs(colour = "") +
theme(legend.position="top") +
theme(legend.text=element_text(size=10));
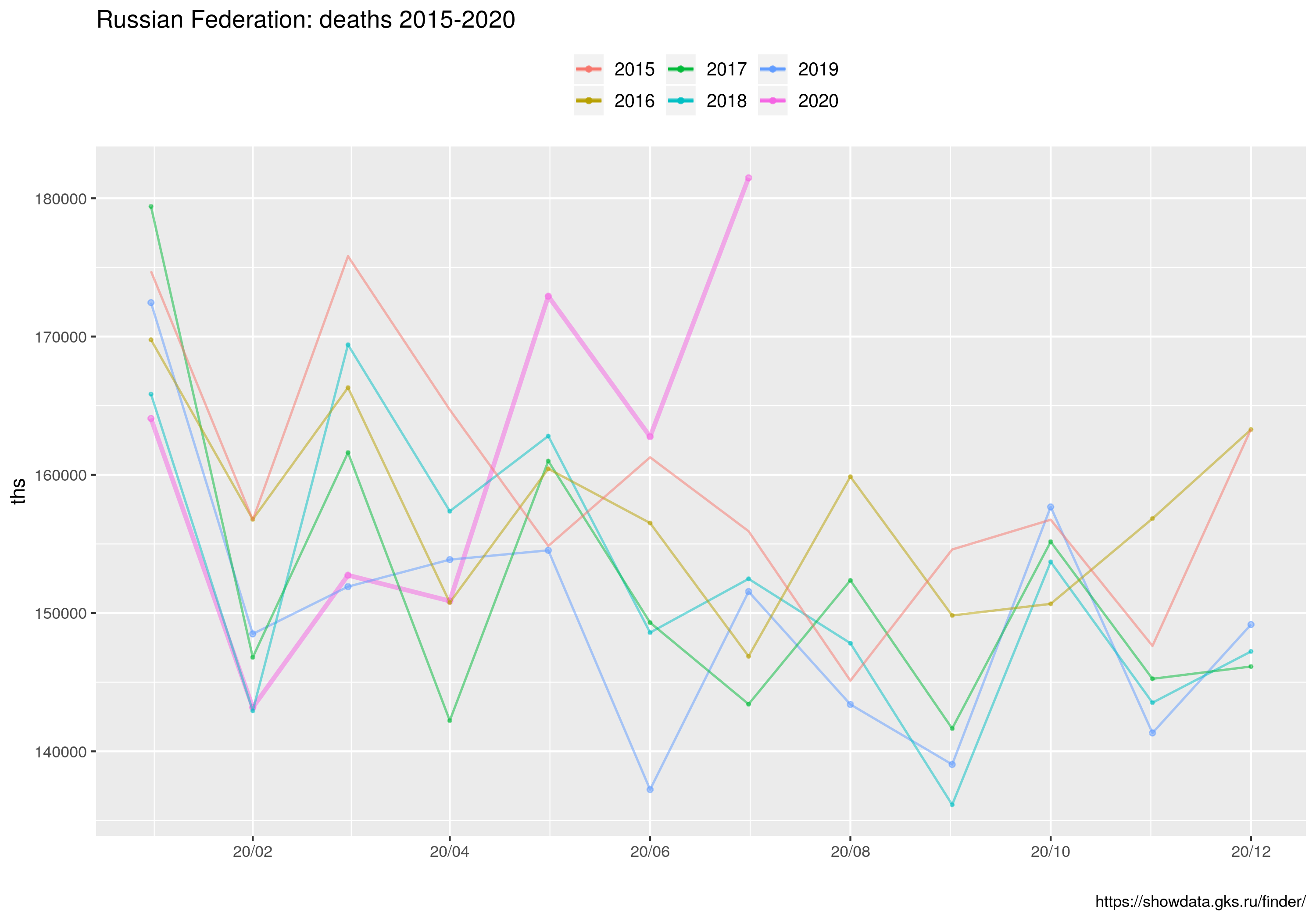
qf <- ggplot() +
ggtitle("Russian Federation: deaths 2015-2020") +
geom_line( data = tt.yy2020, mapping = aes(x=yyyymmdd, y = zgony, colour = '2020'), alpha=.5, size=size0) +
geom_point( data = tt.yy2020, mapping = aes(x=yyyymmdd, y = zgony, colour = '2020'), alpha=.5, size=size0) +
geom_line( data = tt.yy2019, mapping = aes(x=yyyymmdd, y = zgony, colour = '2019'), alpha=.5, size=size1) +
geom_point( data = tt.yy2019, mapping = aes(x=yyyymmdd, y = zgony, colour = '2019'), alpha=.5, size=size0) +
geom_line( data = tt.yy2018, mapping = aes(x=yyyymmdd, y = zgony, colour = '2018'), alpha=.5, size=size1) +
geom_point( data = tt.yy2018, mapping = aes(x=yyyymmdd, y = zgony, colour = '2018'), alpha=.5, size=size1) +
geom_line( data = tt.yy2017, mapping = aes(x=yyyymmdd, y = zgony, colour = '2017'), alpha=.5, size=size1) +
geom_point( data = tt.yy2017, mapping = aes(x=yyyymmdd, y = zgony, colour = '2017'), alpha=.5, size=size1) +
###
geom_line( data = tt.yy2016, mapping = aes(x=yyyymmdd, y = zgony, colour = '2016'), alpha=.5, size=size1) +
geom_point( data = tt.yy2016, mapping = aes(x=yyyymmdd, y = zgony, colour = '2016'), alpha=.5, size=size1) +
geom_line( data = tt.yy2015, mapping = aes(x=yyyymmdd, y = zgony, colour = '2015'), alpha=.5, size=size2) +
geom_point( data = tt.yy2015, mapping = aes(x=yyyymmdd, y = zgony, colour = '2015'), alpha=.5, size=size2) +
scale_x_date( labels = date_format("%y/%m"), breaks = "2 months") +
labs(caption="https://showdata.gks.ru/finder/") +
ylab(label="ths") +
xlab(label="") +
labs(colour = "") +
theme(legend.position="top") +
theme(legend.text=element_text(size=10));
qf <- ggplot() +
ggtitle("Russian Federation: natural balance 2015-2020") +
geom_line( data = tt.yy2020, mapping = aes(x=yyyymmdd, y = diff, colour = '2020'), alpha=.5, size=size0) +
geom_point( data = tt.yy2020, mapping = aes(x=yyyymmdd, y = diff, colour = '2020'), alpha=.5, size=size0) +
geom_line( data = tt.yy2019, mapping = aes(x=yyyymmdd, y = diff, colour = '2019'), alpha=.5, size=size1) +
geom_point( data = tt.yy2019, mapping = aes(x=yyyymmdd, y = diff, colour = '2019'), alpha=.5, size=size0) +
geom_line( data = tt.yy2018, mapping = aes(x=yyyymmdd, y = diff, colour = '2018'), alpha=.5, size=size1) +
geom_point( data = tt.yy2018, mapping = aes(x=yyyymmdd, y = diff, colour = '2018'), alpha=.5, size=size1) +
geom_line( data = tt.yy2017, mapping = aes(x=yyyymmdd, y = diff, colour = '2017'), alpha=.5, size=size1) +
geom_point( data = tt.yy2017, mapping = aes(x=yyyymmdd, y = diff, colour = '2017'), alpha=.5, size=size1) +
###
geom_line( data = tt.yy2016, mapping = aes(x=yyyymmdd, y = diff, colour = '2016'), alpha=.5, size=size1) +
geom_point( data = tt.yy2016, mapping = aes(x=yyyymmdd, y = diff, colour = '2016'), alpha=.5, size=size1) +
geom_line( data = tt.yy2015, mapping = aes(x=yyyymmdd, y = diff, colour = '2015'), alpha=.5, size=size2) +
geom_point( data = tt.yy2015, mapping = aes(x=yyyymmdd, y = diff, colour = '2015'), alpha=.5, size=size2) +
scale_x_date( labels = date_format("%y/%m"), breaks = "2 months") +
labs(caption="https://showdata.gks.ru/finder/") +
ylab(label="ths") +
xlab(label="") +
labs(colour = "") +
theme(legend.position="top") +
theme(legend.text=element_text(size=10));
Wreszcie wykres kombinowany liniowo-słupkowy dla miesięcy jednoimiennych (MJ). Linie to 1) średnia liczby zgonów w latach 2015--2020, 2) średnia liczba zgonów w latach 2015--2020 plus/minus odchylenie standardowe liczby zgonów (dla MJ); 3) liczba zgonów w roku 2020. Wykres słupkowy to różnica między liczbą zgonów w roku 2020 a maksymalną liczbą zgonów w latach 2015-2019 ORAZ liczba zgonów z powodu COVID19 (z bazy ECDC). Ten skrypt korzysta z danych z pliku covid_ru.csv, który powstał przez zagregowanie danych dziennych dostępnych ze strony ECDC (dla Federacji Rosyjskiej) oraz policzenie średnich/odchyleń dla jednoimiennych miesięcy z danych w pliku UZRT.csv
library(ggplot2)
library(dplyr)
library(tidyr)
library(scales)
options(dplyr.print_max = 1e9)
#
tt <- read.csv("covid_ru.csv", sep = ';', header=T, na.string="NA");
tt$diff <- tt$urodzenia - tt$zgony
cols <- c("mean19"="navyblue","mean+sd19"="#3591d1", "mean-sd19"="#3591d1",
"deaths20"='brown4', "diff19"="red", "c19d"='cyan')
pf <- ggplot(data=tt) +
ggtitle("Russian Federation: deaths 2015--2020",
subtitle='mean19/sd19: mean/sd 2015--19; diff19: 2020 - max(2015-019); deaths20: deaths 2020; c19d: C19 deaths') +
geom_line(mapping = aes(x=yyyymmdd, y = mean, colour = 'mean19'), alpha=.5, size=.8) +
geom_line(mapping = aes(x=yyyymmdd, y = mean + sd, colour = 'mean+sd19'), alpha=.5, size=.8) +
geom_line(mapping = aes(x=yyyymmdd, y = mean - sd, colour = 'mean-sd19'), alpha=.5, size=.8) +
geom_text(mapping = aes(x=yyyymmdd, label=difflabel, y=diff), vjust=-1.0, size=3 ) +
##geom_line(mapping = aes(x=yyyymmdd, y = c19d, colour = 'c19d'), alpha=.5, size=1.2) +
labs(caption = "Source: https://showdata.gks.ru/finder/; https://www.ecdc.europa.eu/en/covid-19-pandemic") +
##
geom_bar(mapping = aes(x=yyyymmdd, y = diff, fill = 'diff19' ), color='brown', stat="identity", alpha=.25) +
geom_bar(mapping = aes(x=yyyymmdd, y = c19d, fill = 'c19d' ), color ='navyblue', stat="identity", alpha=.25) +
###
geom_line(mapping = aes(x=yyyymmdd, y = deaths, colour = 'deaths20'), alpha=.5, size=1.2) +
scale_x_date( labels = date_format("%y/%m"), breaks = "2 months") +
ylab(label="ths") +
xlab(label="") +
##labs(colour = "Legend") +
scale_colour_manual(name="Lines: ", values=cols) +
scale_fill_manual(name="Bars: ", values=cols) +
theme(legend.position="right") +
theme(legend.text=element_text(size=12));
Skrypt pomocniczy liczący średnie 2015--2019 dla miesięcy jednoimiennych itp rzeczy
library(dplyr)
library(tidyr)
## Agreguje zgodny wg miesięcy jednoimiennych (MJ) dla lat 2015-19
## drukuje średnią/sd/max dla MJ
## Drukuje z2020 - max(z2015--2019)
tt <- read.csv("UZRT.csv", sep = ';', header=T, na.string="NA");
tt$rok <- substr(tt$month, 1, 4)
tt$yyyymmdd <- as.Date(sprintf ("%s-%s", substr(tt$month, 6, 7), substr(tt$month, 9, 10) ), format="%m-%d")
tt$diff <- tt$urodzenia - tt$zgony
tt.yy2020 <- tt %>% filter(rok==2020) %>% as.data.frame
tt.yy2019 <- tt %>% filter(rok==2019) %>% as.data.frame
tt.yy2018 <- tt %>% filter(rok==2018) %>% as.data.frame
tt.yy2017 <- tt %>% filter(rok==2017) %>% as.data.frame
tt.yy2016 <- tt %>% filter(rok==2016) %>% as.data.frame
tt.yy2015 <- tt %>% filter(rok==2015) %>% as.data.frame
## max zgony
tt.yy2020$max.so.far <- pmax (tt.yy2019$zgony, tt.yy2018$zgony,
tt.yy2017$zgony, tt.yy2016$zgony, tt.yy2015$zgony)
## średnia
tt.yy2020$mean <- (tt.yy2019$zgony + tt.yy2018$zgony
+ tt.yy2017$zgony + tt.yy2016$zgony + tt.yy2015$zgony)/5
## odchylenie std:
tt.yy2020$sqmean <- (tt.yy2019$zgony^2 + tt.yy2018$zgony^2 +
tt.yy2017$zgony^2 + tt.yy2016$zgony^2 + tt.yy2015$zgony^2)/5
tt.yy2020$sq <- sqrt(tt.yy2020$sqmean - tt.yy2020$mean^2)
tt.yy2020$diff20 <- tt.yy2020$zgony - tt.yy2020$max.so.far
sprintf ("%s;%.2f;%.2f;%.2f;%.2f",
tt.yy2020$yyyymmdd, tt.yy2020$diff20, tt.yy2020$sqmean, tt.yy2020$sq, tt.yy2020$max.so.far )

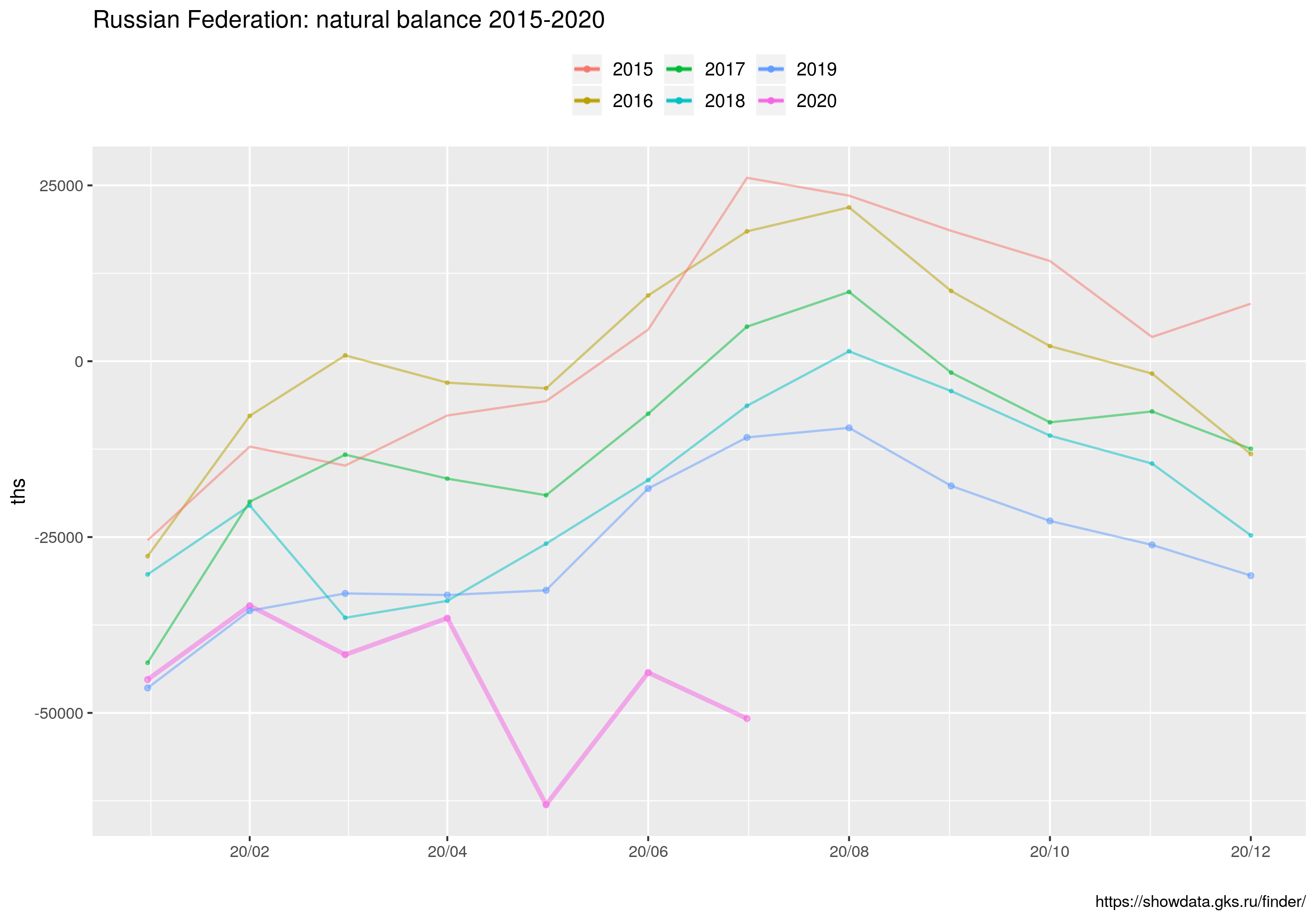
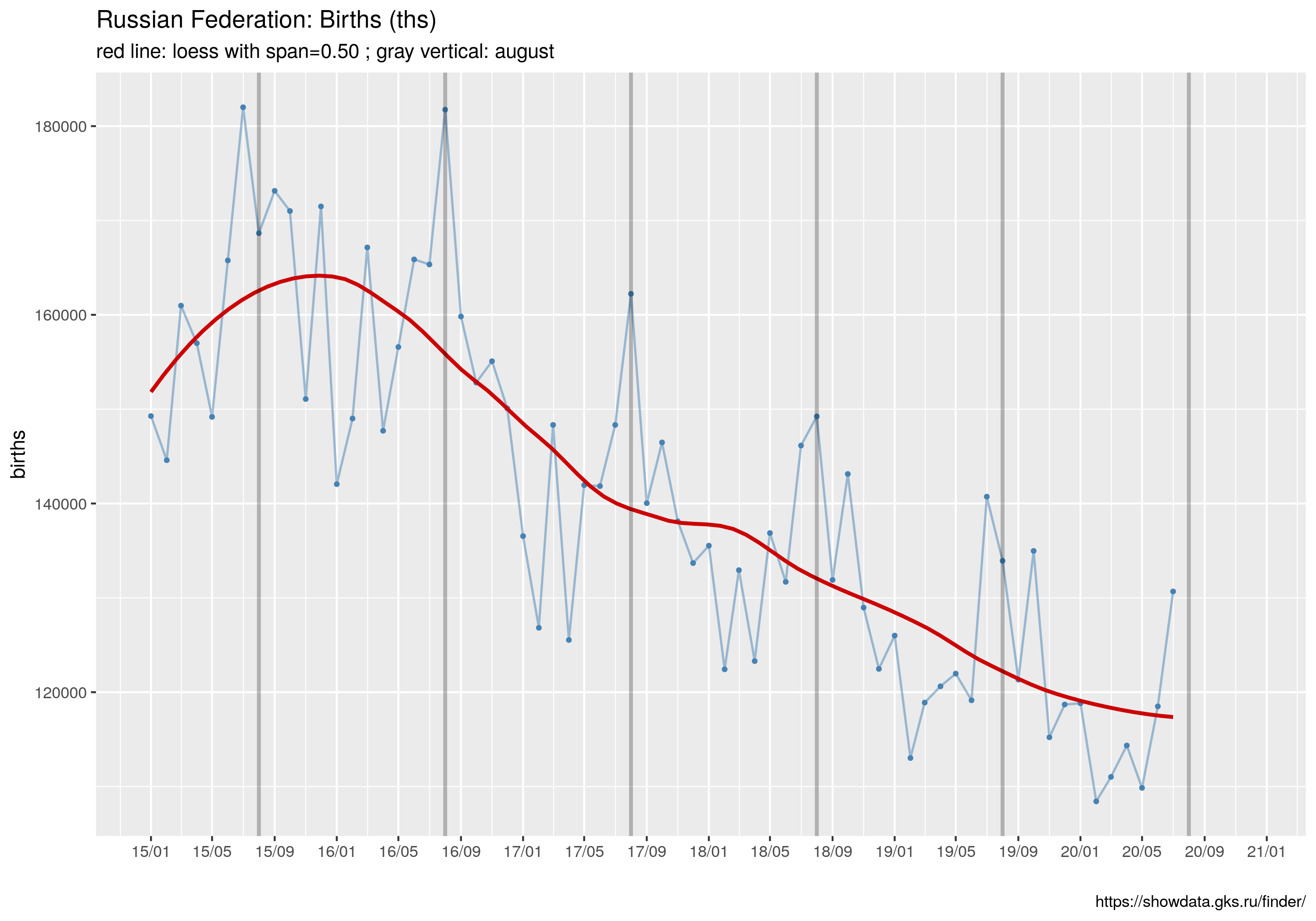
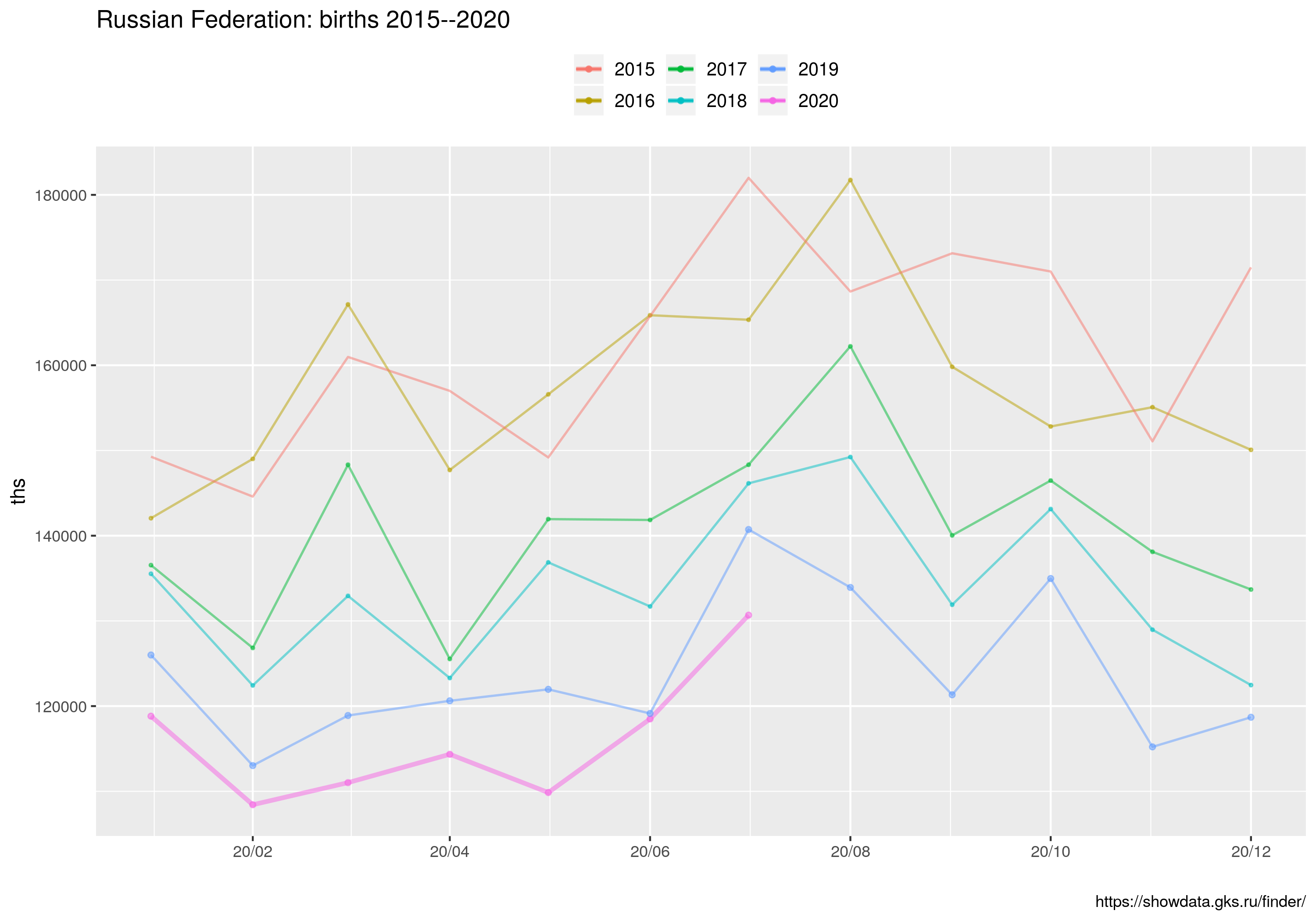
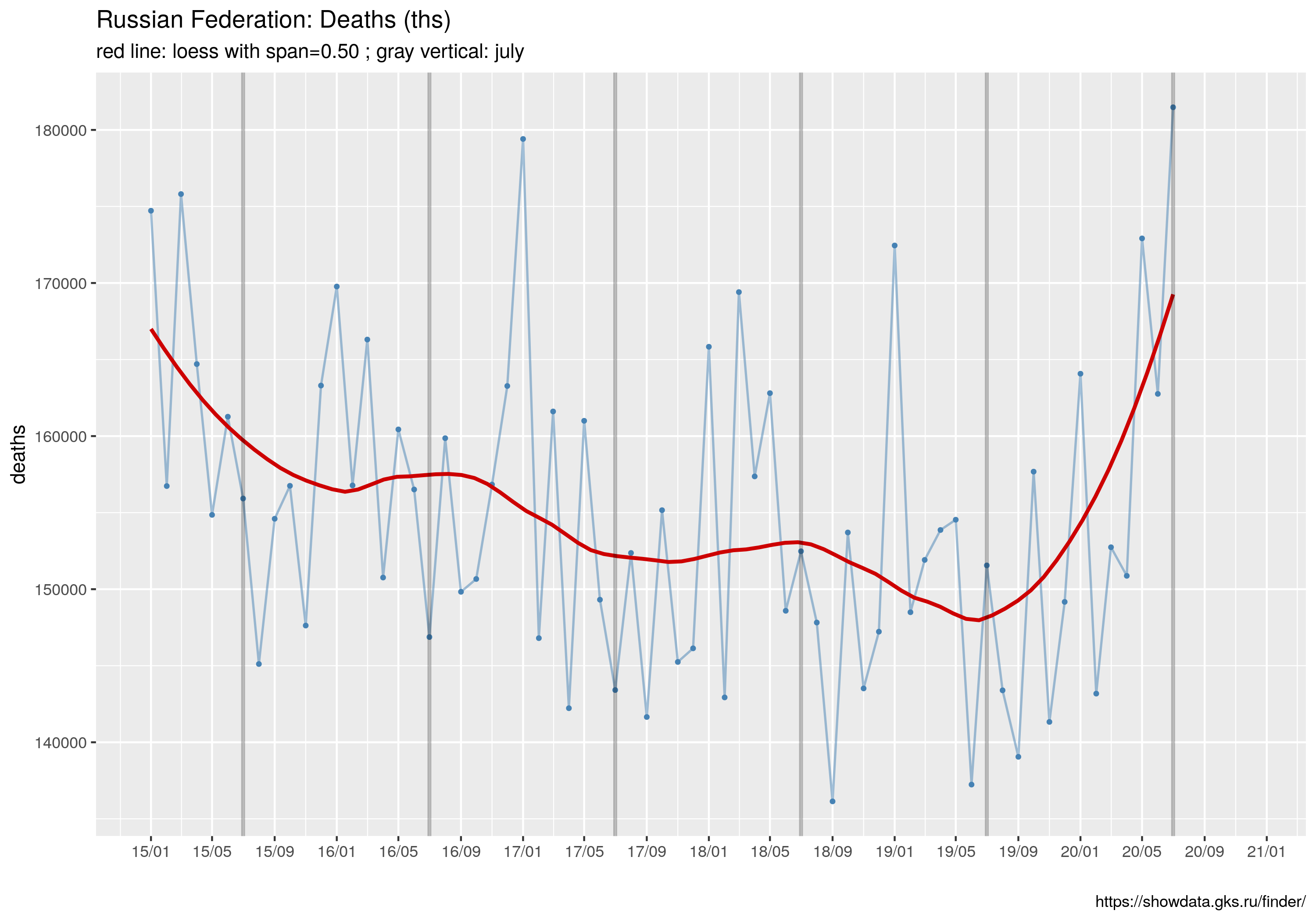
W tym wpisie znajdują się wszystkie najważniejsze informacje.
OdpowiedzUsuń